Back to basics with Pandas
In the spirit of our last post, let’s take things back to basics with a simple end-to-end data analysis example.
Back to basics with pandas, plotnine, and sklearn
In this colab we do a quick review of the ‘data science basics’:
- Download a (small) dataset from an online repository.
- Use
pandasto store and manipulate the data. - Use
plotnineto generate plots/visualizations. - Use
scikit-learnto fit a simple logistic regression model.
# @title Imports
import numpy as np
import pandas as pd
import plotnine as gg
import requests
import sklearn
Let there be data
Let’s fetch the famous Iris dataset from the UCI machine learning datasets repository.
# The URL for the raw data in text form.
data_url = 'https://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data'
# Column names/metadata.
feature_columns = ['sepal_length', 'sepal_width', 'petal_length', 'petal_width']
target_columns = ['target']
# Fetch the raw data from the website.
response = requests.get(url=data_url)
rows = response.text.split('\n')
data = [r.split(',') for r in rows]
# Turn it into a dataframe and drop any null values.
df = pd.DataFrame(data, columns=feature_columns + target_columns)
df.dropna(inplace=True)
# Fix up the data types: features are numeric, targets are categorical.
df[feature_columns] = df[feature_columns].astype('float')
df[target_column] = df[target_column].astype('category')
Take a quick look at the data
# Look at the raw data to check it looks sensible.
df.head()
| sepal_length | sepal_width | petal_length | petal_width | target | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
# Look at some summary statistics.
df.describe()
| sepal_length | sepal_width | petal_length | petal_width | |
|---|---|---|---|---|
| count | 150.000000 | 150.000000 | 150.000000 | 150.000000 |
| mean | 5.843333 | 3.054000 | 3.758667 | 1.198667 |
| std | 0.828066 | 0.433594 | 1.764420 | 0.763161 |
| min | 4.300000 | 2.000000 | 1.000000 | 0.100000 |
| 25% | 5.100000 | 2.800000 | 1.600000 | 0.300000 |
| 50% | 5.800000 | 3.000000 | 4.350000 | 1.300000 |
| 75% | 6.400000 | 3.300000 | 5.100000 | 1.800000 |
| max | 7.900000 | 4.400000 | 6.900000 | 2.500000 |
# 6.1 KB ... now that's some #bigdata ;)
df.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 150 entries, 0 to 149
Data columns (total 5 columns):
sepal_length 150 non-null float64
sepal_width 150 non-null float64
petal_length 150 non-null float64
petal_width 150 non-null float64
target 150 non-null category
dtypes: category(1), float64(4)
memory usage: 6.1 KB
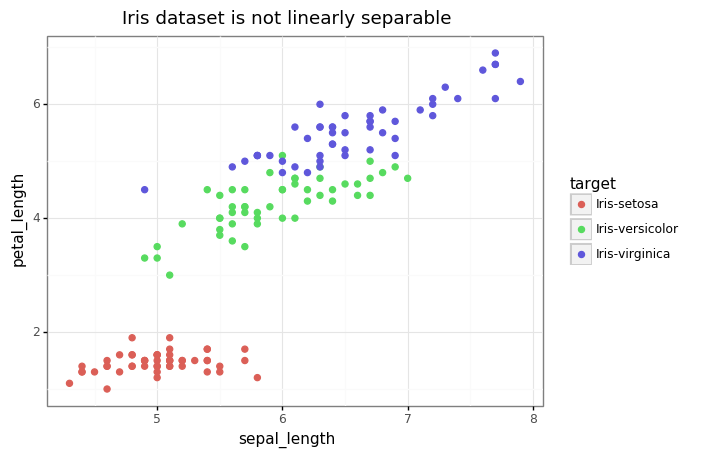
# Let's take a visual look at the data now, projecting down to 2 dimensions.
gg.theme_set(gg.theme_bw())
p = (gg.ggplot(df)
+ gg.aes(x='sepal_length', y='petal_length', color='target')
+ gg.geom_point(size=2)
+ gg.ggtitle('Iris dataset is not linearly separable')
)
p
Fitting a simple linear model
Let’s get ‘old school’ and fit a simple logistic regression classifier using the raw features.
In the binary case, this is a linear probabilistic model with Bernoulli likelihood given by
\[\mathcal{L}\left(\pmb{x}, y \lvert \pmb{x}\right) = f(\pmb{x})^y (1 - f(\pmb{x}))^{1-y},\]where \(f(\pmb{x}) = \sigma\left(\pmb{w}^T\pmb{x}\right)\).
Assuming the dataset \(\mathcal{D}=\left\{\pmb{x}^{(i)}, y^{(i)}\right\}_{i=1}^{n}\) is iid, the negative log-likelihood yields the familiar cross-entropy loss:
\[\begin{aligned} \log\mathcal{L}\left(\mathcal{D} \lvert \pmb{w}\right) &= \log\prod_{i=1}^{n} \sigma\left(\pmb{w}^T\pmb{x}^{(i)}\right)^{y^{(i)}} \left(1 - \sigma\left(\pmb{w}^T\pmb{x}^{(i)}\right)\right)^{1-y^{(i)}} \\ &= \sum_{i=1}^n y^{(i)}\log f\left(\pmb{x}^{(i)}\right) + (1-y^{(i)})\log\left(1 - f\left(\pmb{x}^{(i)}\right)\right) \end{aligned}\]This is a convex and trivially differnetiable objective that we can optimize with gradient-based methods. We’ll defer this to scikit-learn (which also does some other tricks + regularization).
# Create a logistic regression model.
model = sklearn.linear_model.LogisticRegression(solver='lbfgs',
multi_class='multinomial')
# Extract the features and targets as numpy arrays.
X = df[feature_columns].values
y = df[target_column].cat.codes # Note that we 'code' the categories as integers.
# Fit the model.
model.fit(X, y)
# Compute the model accuracy over the training set (we don't have a test set).
pred_df = df.copy()
pred_df['prediction'] = model.predict(X).astype('int8')
pred_df['correct'] = pred_df.prediction == pred_df.target.cat.codes
accuracy = pred_df.correct.sum() / len(pred_df)
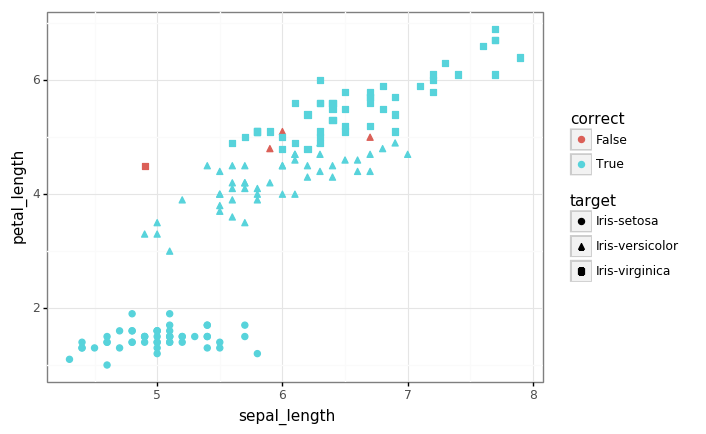
print('Accuracy: {:.3f}'.format(accuracy))
Accuracy: 0.973
# Let's look again at the data, highlighting the examples which we mis-classified.
p = (gg.ggplot(pred_df)
+ gg.aes(x='sepal_length', y='petal_length', shape='target', color='correct')
+ gg.geom_point(size=2)
)
p